For more on our big picture scientific questions, including publications sorted by topic, please visit the research page.
UW lab members in bold
For the most up-to-date list, see Google Scholar
(20) Danielle M Stevens, Alba Moreno-Pérez, Alexandra J Weisberg, Charis Ramsing, Judith Fliegmann, Ning Zhang, Melanie Madrigal, Gregory Martin, Adam Steinbrenner, Georg Felix, Gitta Coaker. (2023). Evolutionary dynamics of proteinaceous MAMPs reveals intrabacterial antagonism of plant immune perception. bioRxiv. DOI: 10.1101/2023.09.21.558511 (link)
(19) Snoeck S, Garcia AGK, Steinbrenner AD. (2023). Plant Receptor-like proteins (RLPs): Structural features enabling versatile immune recognition. Physiological and Molecular Plant Pathology. DOI: 10.1016/j.pmpp.2023.102004 (link) (pdf)
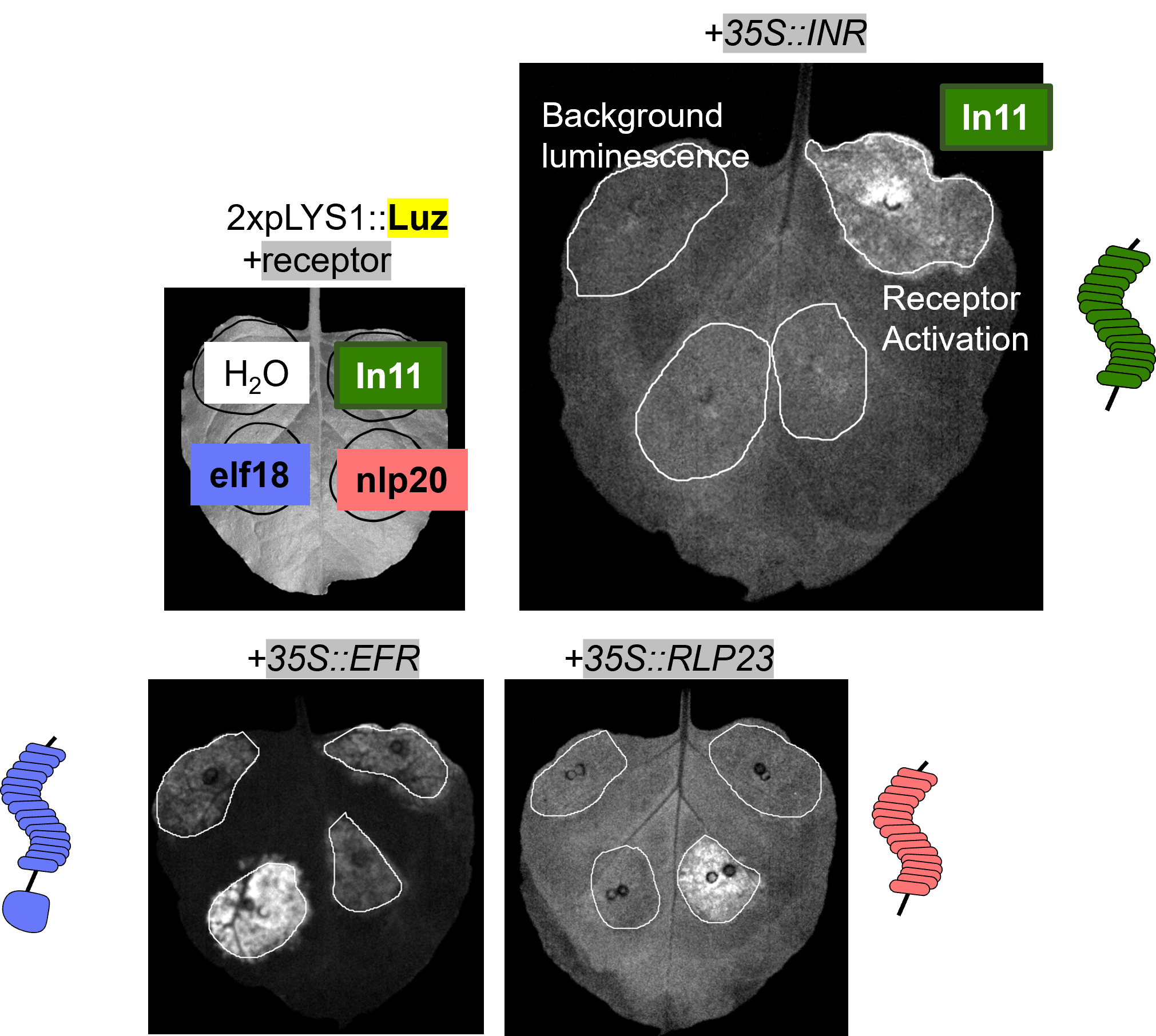
(18) Garcia, AGK, Steinbrenner AD. (2022). Bringing Plant Immunity to Light: A Genetically Encoded, Bioluminescent Reporter of Pattern Triggered Immunity in Nicotiana benthamiana. Molecular Plant-Microbe Interactions. (link) (pdf)
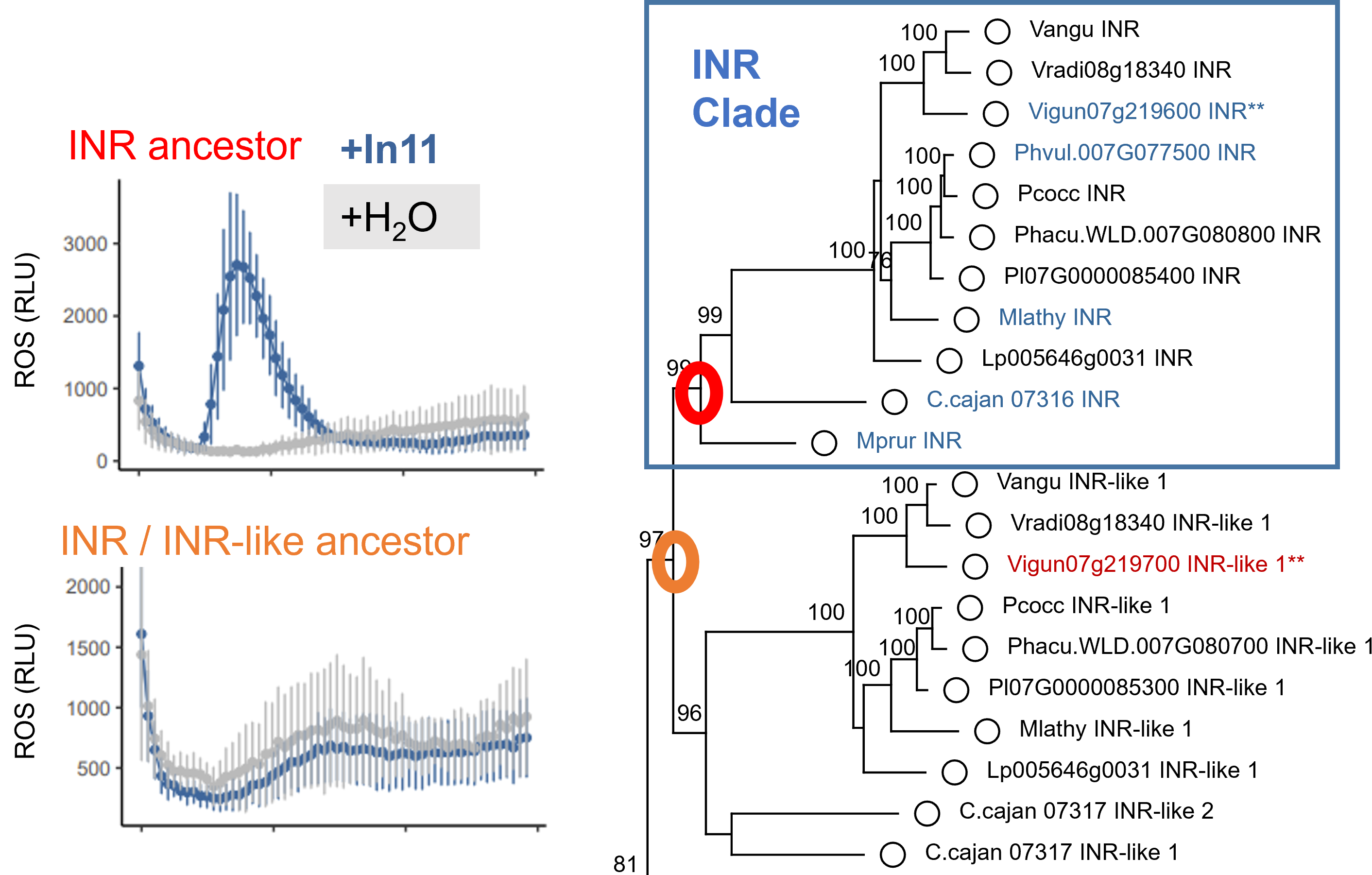
(17) Snoeck S, Abramson BW, Garcia AGK, Egan AN, Michael TP, Steinbrenner AD. (2022). Evolutionary gain and loss of a plant pattern-recognition receptor for HAMP recognition. eLIFE. DOI: 10.7554/eLife.81050 (link) (pdf)
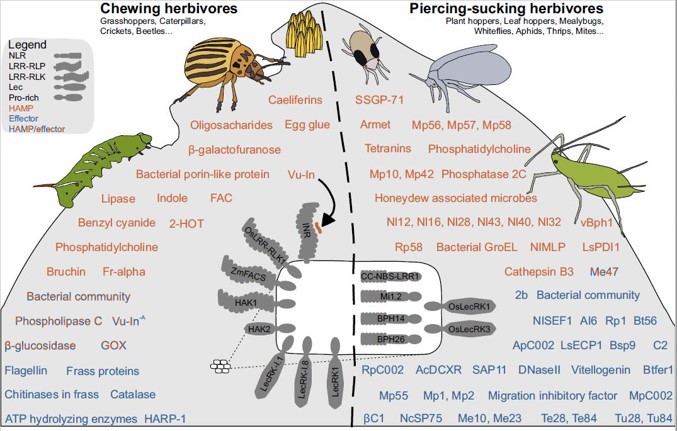
(16) Steinbrenner AD, Saldivar E, Hodges N, Chaparro AF, Guayazán-Palacios N, Schmelz EA. (2021). Signatures of plant defense response specificity mediated by herbivore-associated molecular patterns in legumes. The Plant Journal. DOI: 10.1111/tpj.15732. (link) (pdf)
(15) Snoeck S, Guayazán-Palacios N, Steinbrenner AD. (2022) Molecular tug-of-war: Plant immune recognition of herbivory. The Plant Cell DOI: 10.1093/plcell/koac009. (link) (pdf)
(14) Poretsky E, Ruiz M, Ahmadian N, Steinbrenner AD, Dressano K, Schmelz EA, Huffaker A. (2021). Comparative analyses of responses to exogenous and endogenous antiherbivore elicitors enables a forward genetics approach to identify maize gene candidates mediating sensitivity to Herbivore Associated Molecular Patterns. Plant J. DOI:10.1111/tpj.15510 (link) (pdf)
(13) Bogdan P, Caetano-Anollés G, Jolles A, Kim H, Morris J, Murphy C, Royer C, Snell EH, Steinbrenner AD, Strausfeld N. (2021) Biological networks across scales.Integrative and Comparative Biol. DOI:10.1093/icb/icab069 (link)
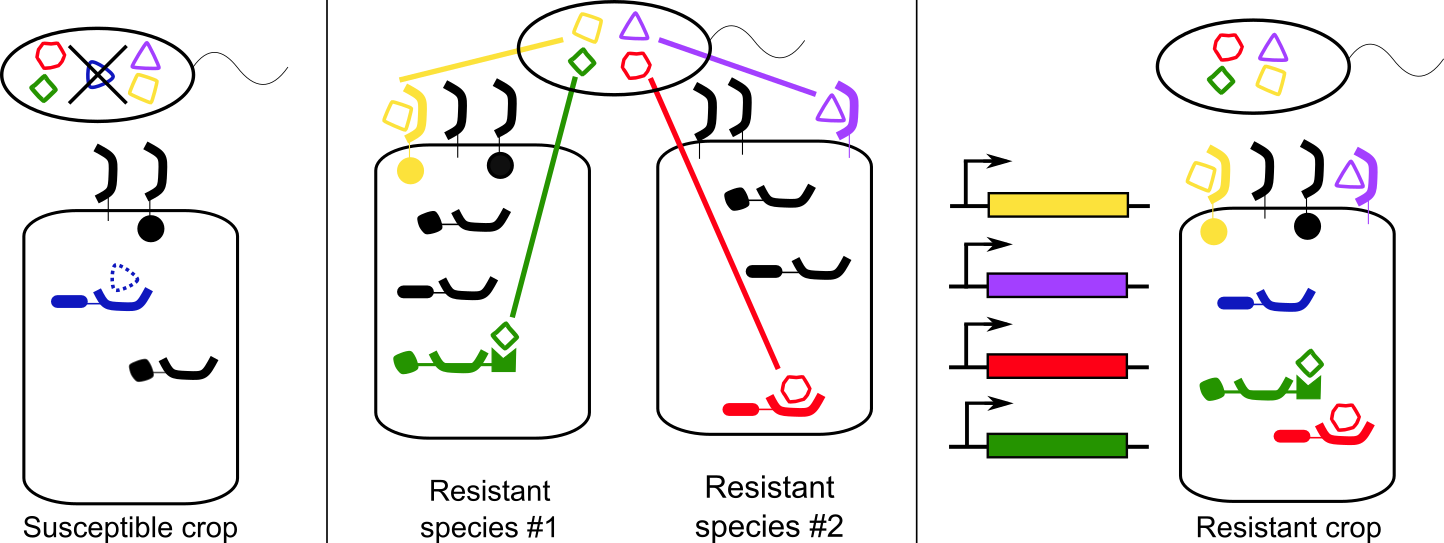
(12) Schultink, A, Steinbrenner, A.D. 2021. A playbook for developing disease-resistant crops through immune receptor identification and transfer. Curr. Opin. Plant Biol. DOI:10.1016/j.pbi.2021.102089 (link) (pdf)
(11) Steinbrenner AD, Maria Muñoz-Amatriaín, Chaparro AF, Aguilar-Venegas JMA, Lo S, Okuda S, Glauser G, Dongiovanni J, Shi D, Hall M, Crubaugh D, Holton N, Zipfel C, Abagyan R, Turlings TCJ, Close TJ, Huffaker A, Schmelz EA. 2020. A receptor-like protein mediates plant immune responses to herbivore-associated molecular patterns. Proc. Nat. Acad. Sci. DOI:10.1073/pnas.2018415117 (link) (pdf)
(10) Steinbrenner AD, 2020. The evolving landscape of cell surface pattern recognition across plant immune networks. Curr. Opin. Plant Biol. DOI: 10.1016/j.pbi.2020.05.001 (link) (pdf)
(9) Schultink A, Qi T, Lee A, Steinbrenner AD, Staskawicz, B. 2017. Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J. DOI: 10.1111/tpj.13715 (link) (pdf)
(8) Goritschnig S*,Steinbrenner AD*, Grunwald DJ, Staskawicz, BJ. 2016.Structurally Distinct Arabidopsis thaliana NLR immune receptors recognize tandem WY domains of an oomycete effector. New Phytologist. DOI: 10.1111/nph.13823 (link) (pdf) *contributed equally
(7) Steinbrenner AD, Goritschnig S, Staskawicz BJ. 2015. Recognition and Activation Domains Contribute to Allele-Specific Responses of an Arabidopsis NLR Receptor to an Oomycete Effector Protein. PLoS Pathogens. DOI: 10.1371/journal.ppat.1004665 (link) (pdf)
(6) Zhang R, Li Y, Cowley TJ, Steinbrenner A, Phillips JM, Yount BL, Baric RS, Weiss, SR. 2015. The nsp1, nsp13, and M proteins contribute to the hepatotropism of murine coronavirus JHM.WU. J. Virol. DOI: 10.1128/JVI.03535-14. (link)
(5) Steinbrenner AD, Goritschnig S, Krasileva KV, Schreiber KJ, Staskawicz BJ. 2012. Effector Recognition and Activation of the Arabidopsis thaliana NLR Innate Immune Receptors. Cold Spring Harbor Symposia on Quant. Biol. 77:249-257. (pdf)
(4) Steinbrenner AD, Agerbirk N, Orians CM, Chew FS 2012. Transient abiotic stresses lead to latent defense and reproductive responses over Brassica rapa life cycle. Chemoecology. 22:239-250. (pdf)
(3) Gómez S,Steinbrenner AD, Osorio S, Schueller M, Ferrieri RA, Fernie AR, Orians CM. 2012. From shoots to roots: transport and metabolic changes in tomato after feeding by a specialist lepidopteran. Entomologia Experimentalis et Applicata. 144:101-111. (pdf)
(2) Steinbrenner AD, Gómez S, et al. 2011. Herbivore-induced changes in tomato primary metabolism: A whole plant perspective. J. Chem. Ecol. 37(12):1294-303. (pdf)
(1) Chou S, Krasileva KV, Holton JM, Steinbrenner AD, Alber T, Staskawicz BJ. 2011. The Hyaloperonospora arabidopsidis ATR1 effector is a repeat protein with distributed recognition surfaces. Proc. Nat. Acad. Sci. 108(32):13323-13328. (pdf)
(20) Danielle M Stevens, Alba Moreno-Pérez, Alexandra J Weisberg, Charis Ramsing, Judith Fliegmann, Ning Zhang, Melanie Madrigal, Gregory Martin, Adam Steinbrenner, Georg Felix, Gitta Coaker. (2023). Evolutionary dynamics of proteinaceous MAMPs reveals intrabacterial antagonism of plant immune perception. bioRxiv. DOI: 10.1101/2023.09.21.558511 (link)
(19) Snoeck S, Garcia AGK, Steinbrenner AD. (2023). Plant Receptor-like proteins (RLPs): Structural features enabling versatile immune recognition. Physiological and Molecular Plant Pathology. DOI: 10.1016/j.pmpp.2023.102004 (link) (pdf)
(18) Garcia, AGK, Steinbrenner AD. (2022). Bringing Plant Immunity to Light: A Genetically Encoded, Bioluminescent Reporter of Pattern Triggered Immunity in Nicotiana benthamiana. Molecular Plant-Microbe Interactions. (link) (pdf)
(17) Snoeck S, Abramson BW, Garcia AGK, Egan AN, Michael TP, Steinbrenner AD. (2022). Evolutionary gain and loss of a plant pattern-recognition receptor for HAMP recognition. eLIFE. DOI: 10.7554/eLife.81050 (link) (pdf)
(16) Steinbrenner AD, Saldivar E, Hodges N, Chaparro AF, Guayazán-Palacios N, Schmelz EA. (2021). Signatures of plant defense response specificity mediated by herbivore-associated molecular patterns in legumes. The Plant Journal. DOI: 10.1111/tpj.15732. (link) (pdf)
(15) Snoeck S, Guayazán-Palacios N, Steinbrenner AD. (2022) Molecular tug-of-war: Plant immune recognition of herbivory. The Plant Cell DOI: 10.1093/plcell/koac009. (link) (pdf)
(14) Poretsky E, Ruiz M, Ahmadian N, Steinbrenner AD, Dressano K, Schmelz EA, Huffaker A. (2021). Comparative analyses of responses to exogenous and endogenous antiherbivore elicitors enables a forward genetics approach to identify maize gene candidates mediating sensitivity to Herbivore Associated Molecular Patterns. Plant J. DOI:10.1111/tpj.15510 (link) (pdf)
(13) Bogdan P, Caetano-Anollés G, Jolles A, Kim H, Morris J, Murphy C, Royer C, Snell EH, Steinbrenner AD, Strausfeld N. (2021) Biological networks across scales.Integrative and Comparative Biol. DOI:10.1093/icb/icab069 (link)
(12) Schultink, A, Steinbrenner, A.D. 2021. A playbook for developing disease-resistant crops through immune receptor identification and transfer. Curr. Opin. Plant Biol. DOI:10.1016/j.pbi.2021.102089 (link) (pdf)
(11) Steinbrenner AD, Maria Muñoz-Amatriaín, Chaparro AF, Aguilar-Venegas JMA, Lo S, Okuda S, Glauser G, Dongiovanni J, Shi D, Hall M, Crubaugh D, Holton N, Zipfel C, Abagyan R, Turlings TCJ, Close TJ, Huffaker A, Schmelz EA. 2020. A receptor-like protein mediates plant immune responses to herbivore-associated molecular patterns. Proc. Nat. Acad. Sci. DOI:10.1073/pnas.2018415117 (link) (pdf)
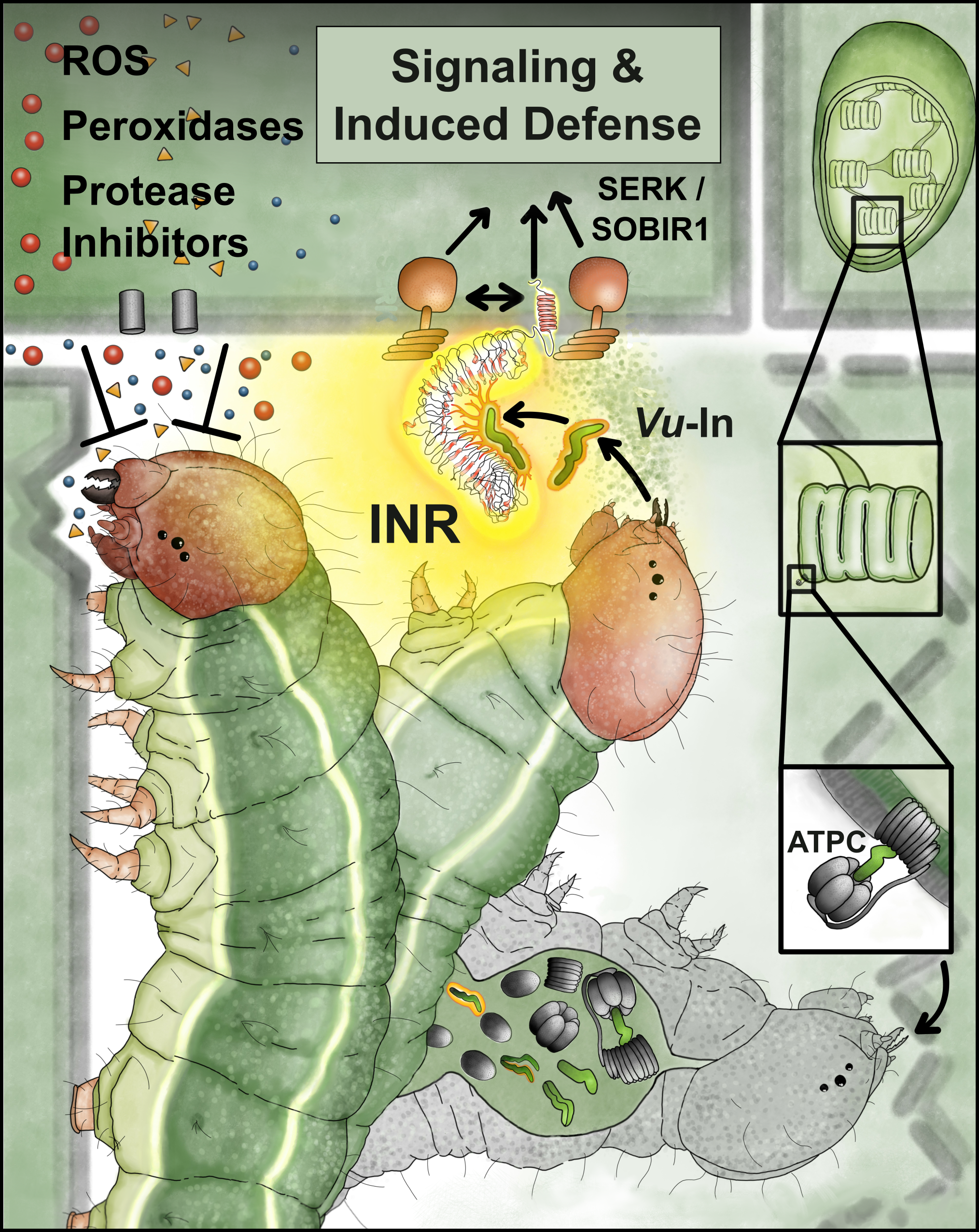
(10) Steinbrenner AD, 2020. The evolving landscape of cell surface pattern recognition across plant immune networks. Curr. Opin. Plant Biol. DOI: 10.1016/j.pbi.2020.05.001 (link) (pdf)
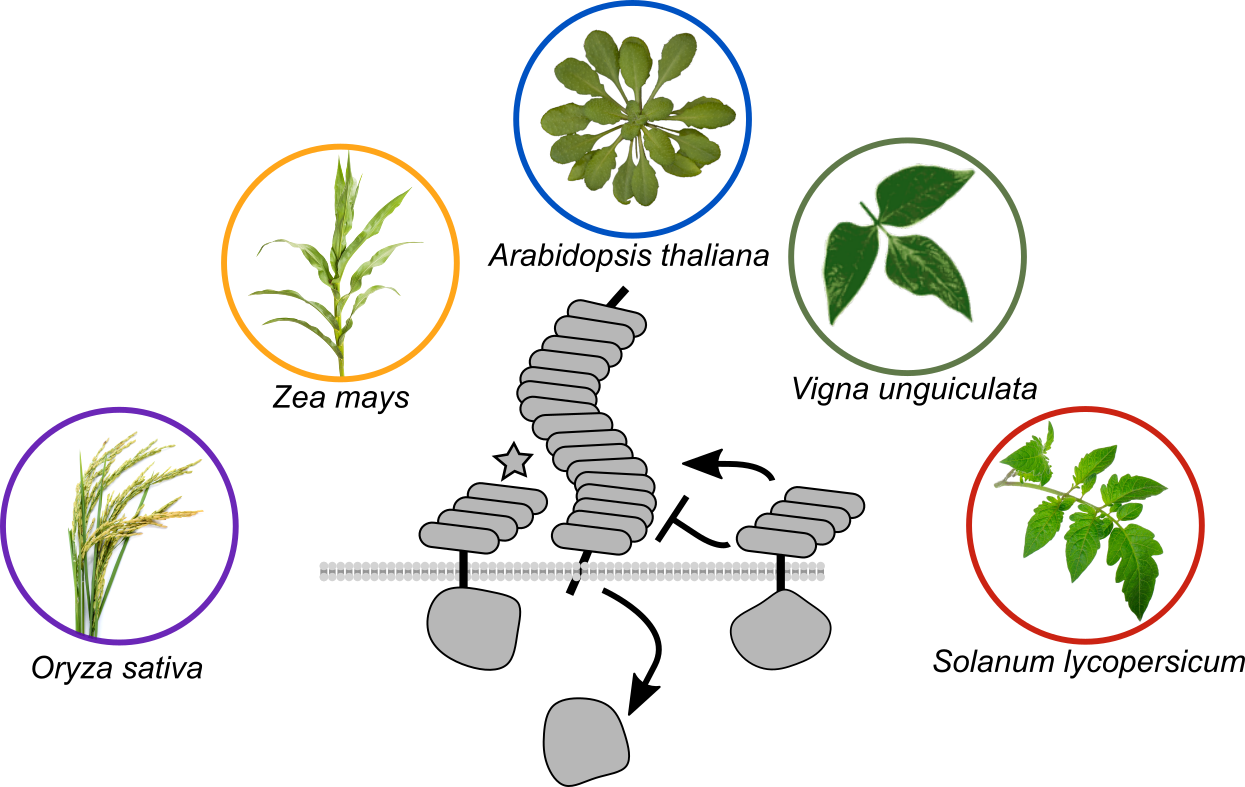
(9) Schultink A, Qi T, Lee A, Steinbrenner AD, Staskawicz, B. 2017. Roq1 mediates recognition of the Xanthomonas and Pseudomonas effector proteins XopQ and HopQ1. Plant J. DOI: 10.1111/tpj.13715 (link) (pdf)
(8) Goritschnig S*,Steinbrenner AD*, Grunwald DJ, Staskawicz, BJ. 2016.Structurally Distinct Arabidopsis thaliana NLR immune receptors recognize tandem WY domains of an oomycete effector. New Phytologist. DOI: 10.1111/nph.13823 (link) (pdf) *contributed equally
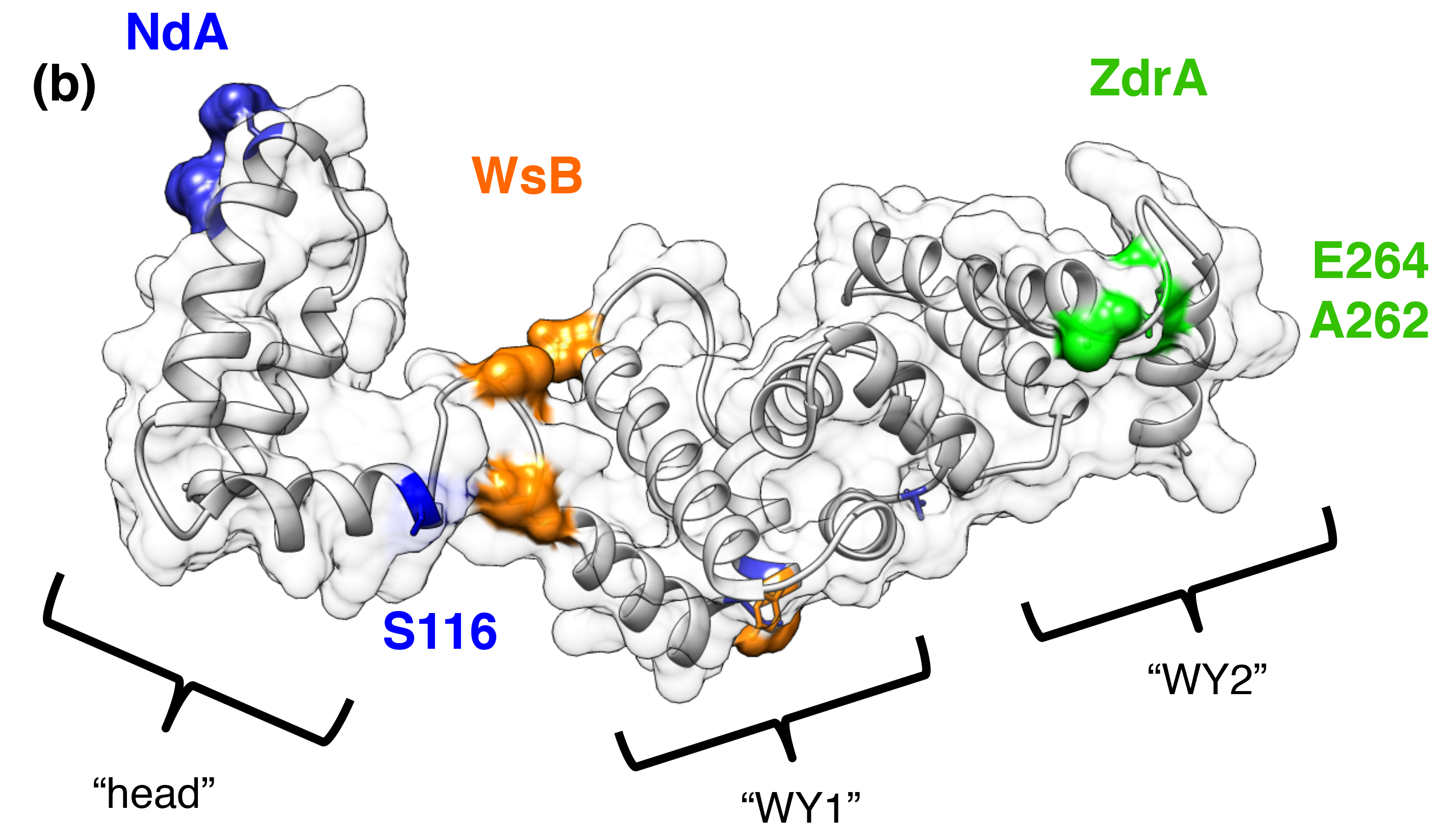
(7) Steinbrenner AD, Goritschnig S, Staskawicz BJ. 2015. Recognition and Activation Domains Contribute to Allele-Specific Responses of an Arabidopsis NLR Receptor to an Oomycete Effector Protein. PLoS Pathogens. DOI: 10.1371/journal.ppat.1004665 (link) (pdf)
(6) Zhang R, Li Y, Cowley TJ, Steinbrenner A, Phillips JM, Yount BL, Baric RS, Weiss, SR. 2015. The nsp1, nsp13, and M proteins contribute to the hepatotropism of murine coronavirus JHM.WU. J. Virol. DOI: 10.1128/JVI.03535-14. (link)
(5) Steinbrenner AD, Goritschnig S, Krasileva KV, Schreiber KJ, Staskawicz BJ. 2012. Effector Recognition and Activation of the Arabidopsis thaliana NLR Innate Immune Receptors. Cold Spring Harbor Symposia on Quant. Biol. 77:249-257. (pdf)
(4) Steinbrenner AD, Agerbirk N, Orians CM, Chew FS 2012. Transient abiotic stresses lead to latent defense and reproductive responses over Brassica rapa life cycle. Chemoecology. 22:239-250. (pdf)
(3) Gómez S,Steinbrenner AD, Osorio S, Schueller M, Ferrieri RA, Fernie AR, Orians CM. 2012. From shoots to roots: transport and metabolic changes in tomato after feeding by a specialist lepidopteran. Entomologia Experimentalis et Applicata. 144:101-111. (pdf)
(2) Steinbrenner AD, Gómez S, et al. 2011. Herbivore-induced changes in tomato primary metabolism: A whole plant perspective. J. Chem. Ecol. 37(12):1294-303. (pdf)
(1) Chou S, Krasileva KV, Holton JM, Steinbrenner AD, Alber T, Staskawicz BJ. 2011. The Hyaloperonospora arabidopsidis ATR1 effector is a repeat protein with distributed recognition surfaces. Proc. Nat. Acad. Sci. 108(32):13323-13328. (pdf)
