Our big questions
-
How do plants first detect attack by chewing insects? We know that plants revamp hormonal and defensive chemistry after attack, but what are the molecular signaling events in the first minutes and seconds after herbivore attack? How are herbivore-specific defenses achieved?
-
How does the plant innate immune system evolve? We focus on cell surface receptors at the plasma membrane, which form a massively diverse gene family in plants underlying pest and pathogen resistance.
1. Plant Immunity
Our lab studies how plants detect diverse attackers. Our lens on this topic is gene families encoding pattern recognition receptors (PRRs) at the plasma membrane, which detect extracellular immunogenic molecular patterns from attackers. Using genetic and genomic tools in both molecular model systems (tobacco, Arabidopsis) and in crop legumes, we can learn which patterns are detected by different plants, and how PRR activation leads to effective resistance against pests and pathogens.
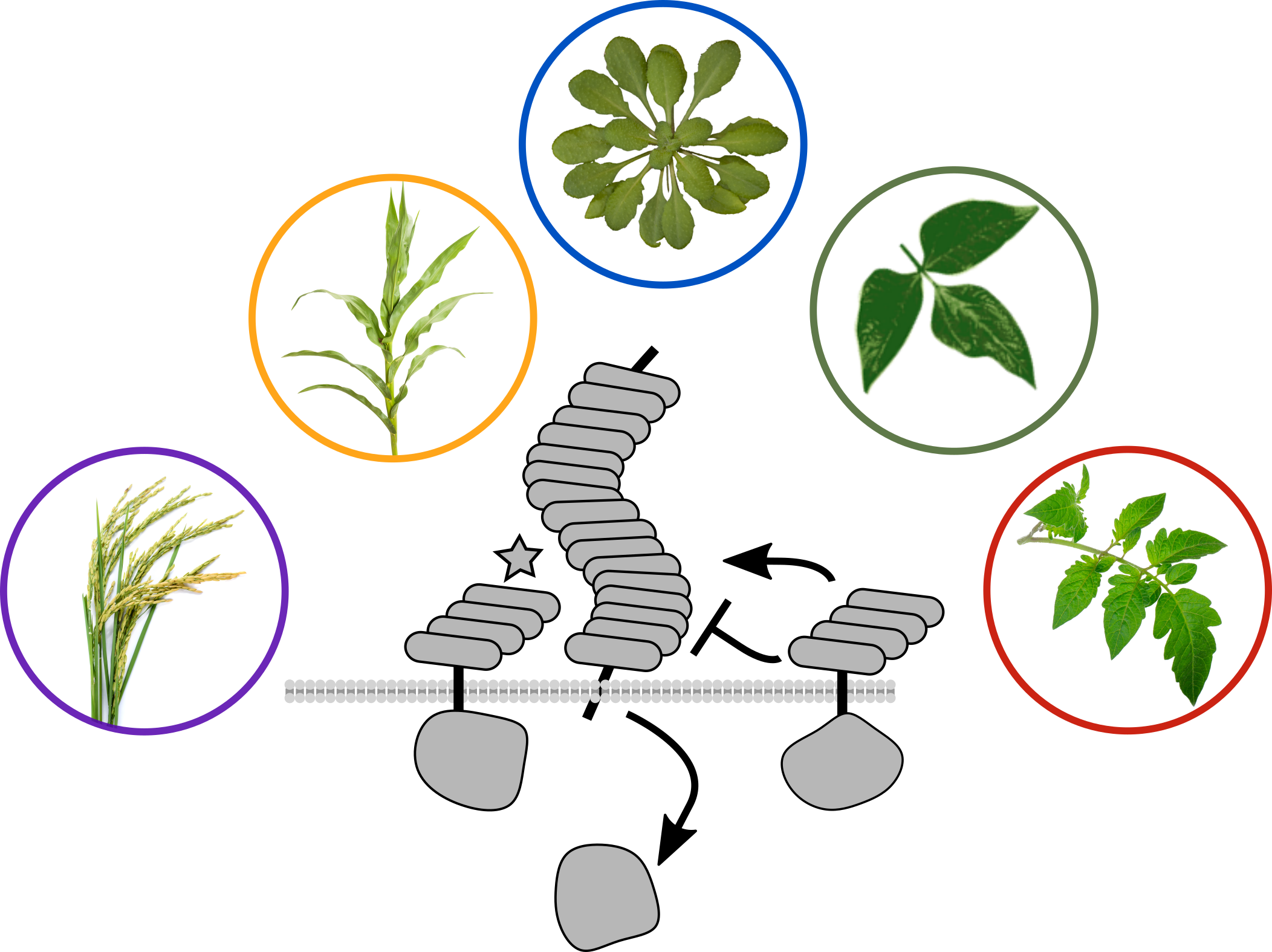
2. Immune recognition of caterpillars
PRRs bind extracellular ligands and initiate defense signal transduction. Many well-characterized receptors detect pathogen-associated molecular patterns (PAMPs), but receptors for analogous herbivore-associated molecular patterns (HAMPs) are not well understood.
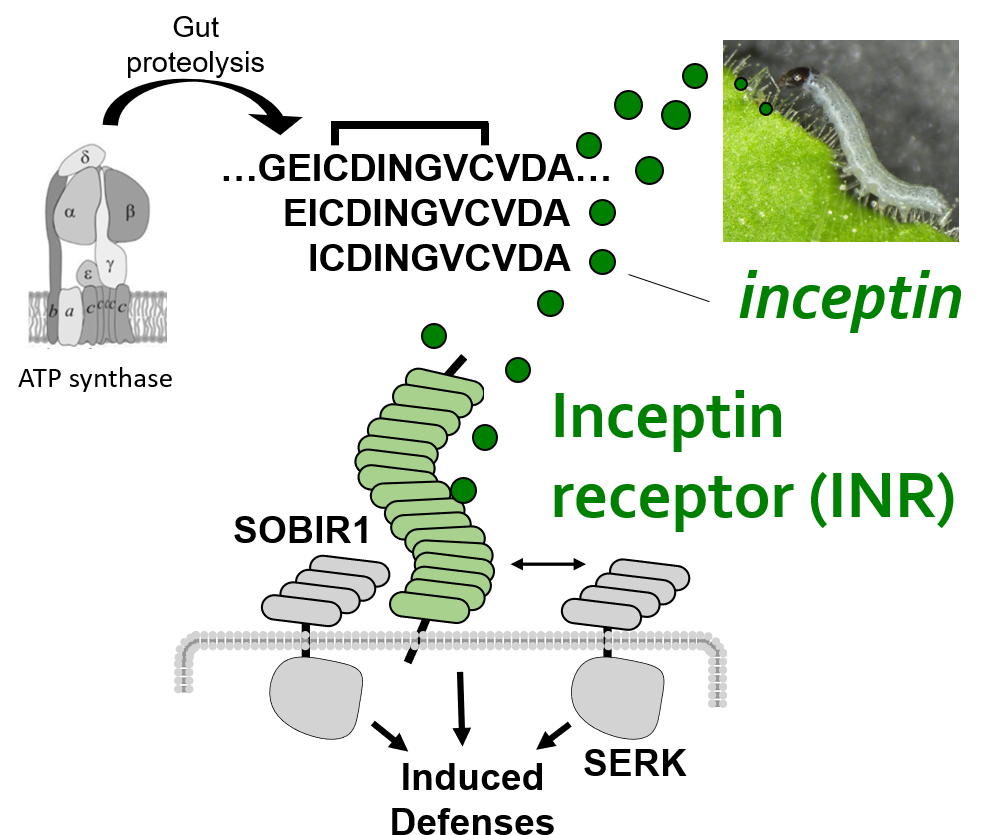
We recently identified a PRR which directly detects a peptide ligand associated with chewing heribivory. Inceptin receptor, or INR, confers binding, signaling, and defensive outputs in response to low nanomolar concentrations of inceptin peptide. We are now using INR to ask how plants activate herbivore-specific defenses, and potential trade-offs with immunity against microbial pathogens.
Our papers on this topic:
- Steinbrenner et al. 2020, “A receptor-like protein mediates plant immune responses to herbivore-associated molecular patterns”, PNAS
- Snoeck et al. 2022, “Molecular tug-of-war: Plant immune recognition of herbivory”, Plant Cell
3. Peptide recognition and differentiation by plant immune receptors
 Plant cell surface receptors signal through a network of co-receptor and adaptor kinases. Canonical receptors recruit the co-receptor BAK1 and kinase cascades lead directly to early immune signaling outputs, such as MAPK phosphorylation, ethylene biosynthesis, and burst of reactive oxygen species.
Plant cell surface receptors signal through a network of co-receptor and adaptor kinases. Canonical receptors recruit the co-receptor BAK1 and kinase cascades lead directly to early immune signaling outputs, such as MAPK phosphorylation, ethylene biosynthesis, and burst of reactive oxygen species.
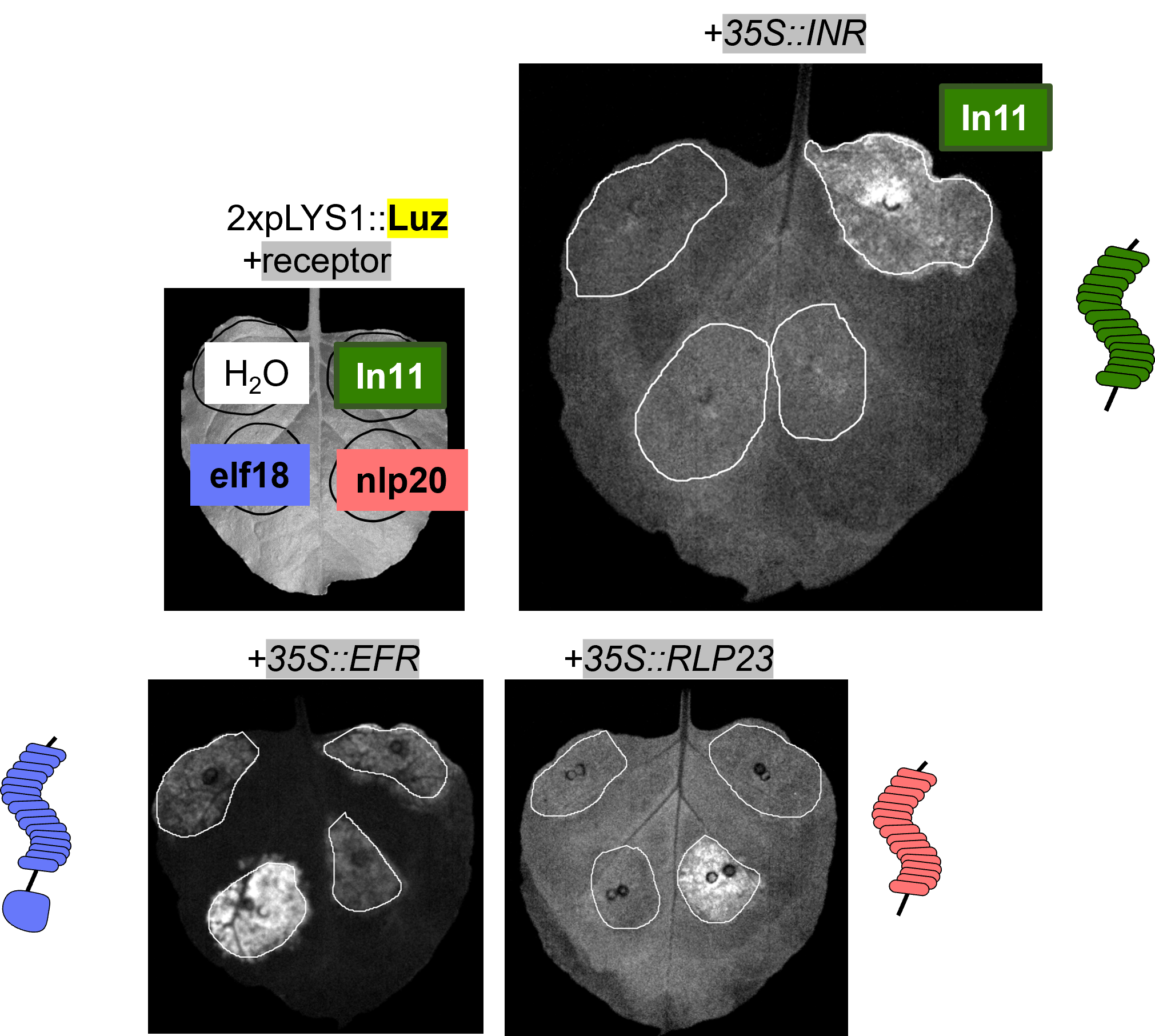
Only a few dozen PRRs have defined ligands, but general principles are emerging regarding ligand specificity encoded by the sequence and structure of LRR ectodomains. We are developing in silico and experimental screening systems to pair receptors and ligands for the vast number of “orphan” receptor sequences annotated across plant genomes.
Our papers on this topic:
- A new tool for studying receptor outputs: Garcia et al. 2022, “Bringing Plant Immunity to Light: A Genetically Encoded, Bioluminescent Reporter of Pattern Triggered Immunity in Nicotiana benthamiana”, bioRxiv
- Steinbrenner 2020, “The evolving landscape of cell surface pattern recognition across plant immune networks”, Curr. Opin. Plant Biol.
4.. Herbivore resistance in legumes
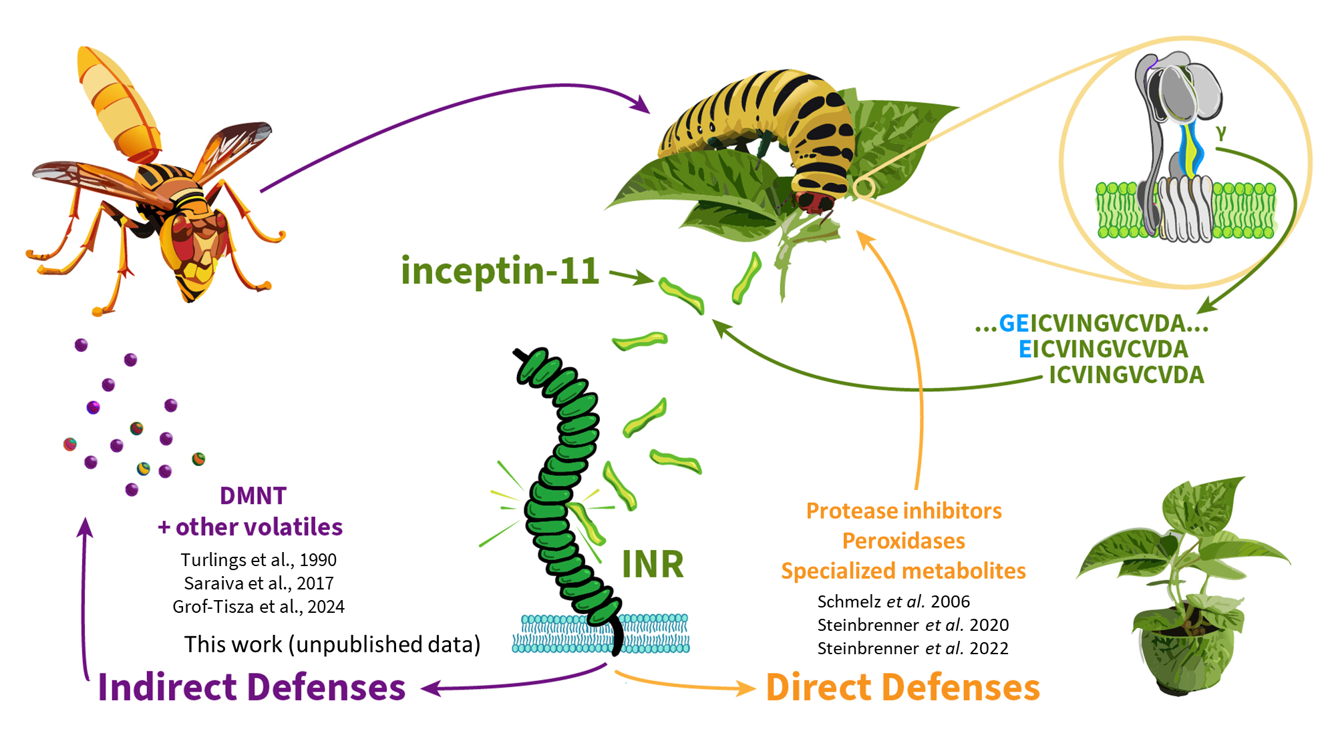 Species in the the legume family (Fabaceae) are uniquely able to respond to inceptins. In cowpea and common bean, inceptin activates canonical anti-herbivore defense responses such as protease inhibition, small molecule toxin biosynthesis, and release of parasitoid-attracting volatiles.
Species in the the legume family (Fabaceae) are uniquely able to respond to inceptins. In cowpea and common bean, inceptin activates canonical anti-herbivore defense responses such as protease inhibition, small molecule toxin biosynthesis, and release of parasitoid-attracting volatiles.
We are interested in the ecological functions of inceptin-triggered responses, especially in relation to the generic wound response. We are developing genetic tools in legumes, including soybean. We are especially interested in how immune receptors allow activation of “indirect defenses” – the recruitment of beneficial ecosystem services.
Our papers on this topic:
- Steinbrenner et al. 2021, “Signatures of plant defense response specificity mediated by herbivore-associated molecular patterns in legumes”, bioRxiv (in press at Plant Journal)
5. Evolution of immune receptor function
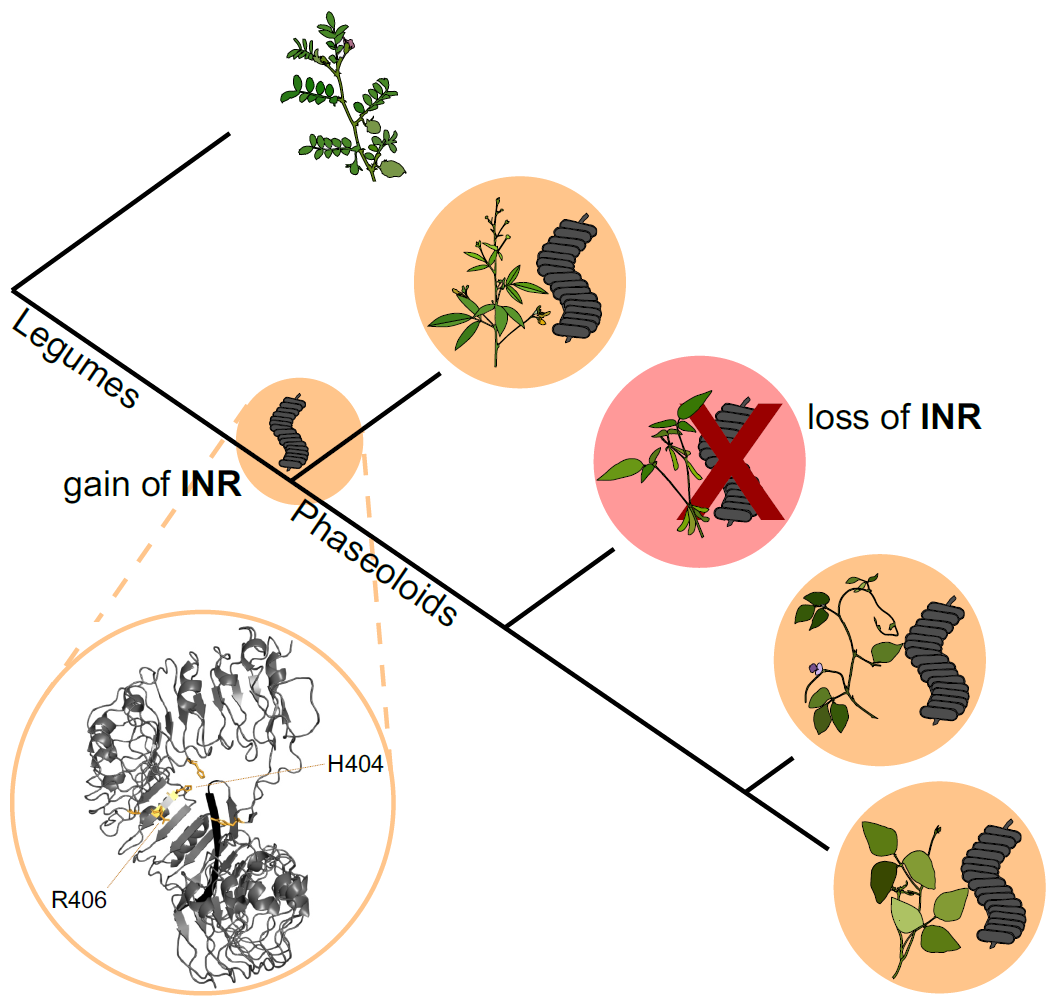 Only a subset of surface receptors are PRRs; others serve critical roles in growth and development. How do new immune functions emerge from ancestral functions? Can we identify useful immune receptors based on genome-level features, especially given the recent deluge of plant pangenomes?
Only a subset of surface receptors are PRRs; others serve critical roles in growth and development. How do new immune functions emerge from ancestral functions? Can we identify useful immune receptors based on genome-level features, especially given the recent deluge of plant pangenomes?
The INR receptor is restricted to certain legume species, but its function is idiosyncratic across species. Soybean, for example, cannot respond to inceptin despite the presence of receptor homologs. We are sequencing additional legume genomes to understand the emergence of INR across 50 million years of legume evolution. We are also transferring INR as a resistance trait to herbivore-susceptible crop species.
Our papers on this topic:
- Snoeck et al. 2022, “Evolutionary gain and loss of a plant pattern-recognition receptor for HAMP recognition”, bioRxiv and in press at ELIFE
- Schultink and Steinbrenner 2021, “A playbook for developing disease-resistant crops through immune receptor identification and transfer”, Curr. Opin. Plant Biol.
- Snoeck et al. 2023, “Plant Receptor-like proteins (RLPs): Structural features enabling versatile immune recognition.” Physiological and Molecular Plant Pathology. (link)
6. Properties and utility of LRR ectodomain networks
In contrast to analogous Toll-like receptors in animals, which contain a structurally similar leucine-rich repeat (LRR) ectodomain, plant immune-related LRR sensors form a highly diversified network of heterotypic ectodomain interactions (Smakowska-Luzan 2018 doi:10.1038/nature25184). We are working to develop orthogonal ligand-sensing systems across plant and non-plant models for biotechnological use in both agriculture and medicine.
